Search options
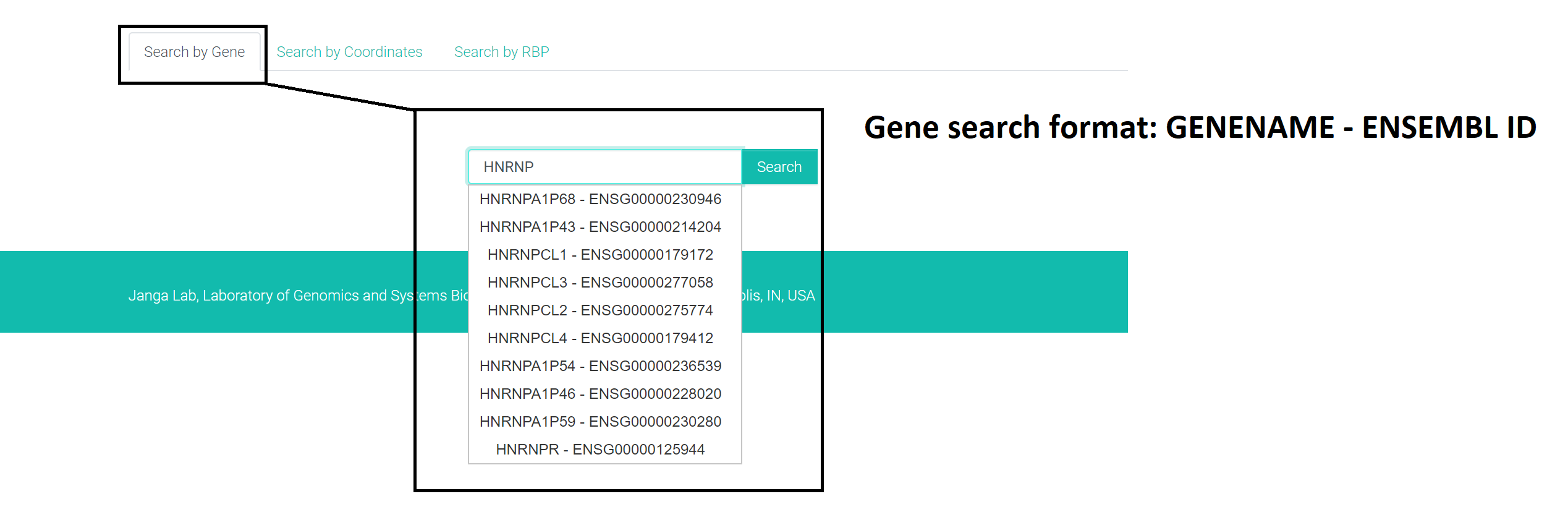
1. Gene based search:
Search format: "GENENAME - ENSEMBLID"
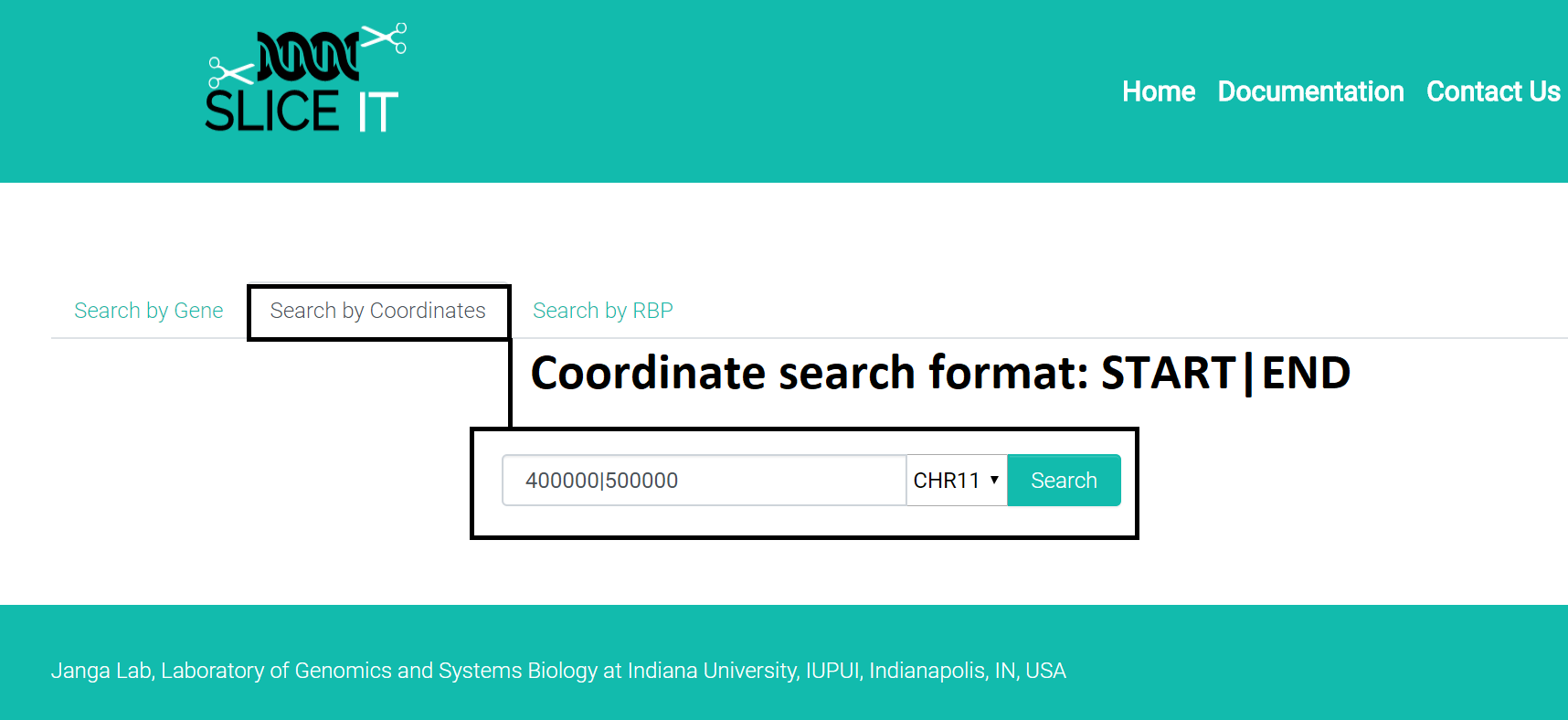
2. Coordinate based search:
Search format: "STARTLOCATION|STOPLOCATION and select the chromosome from the dropdown"
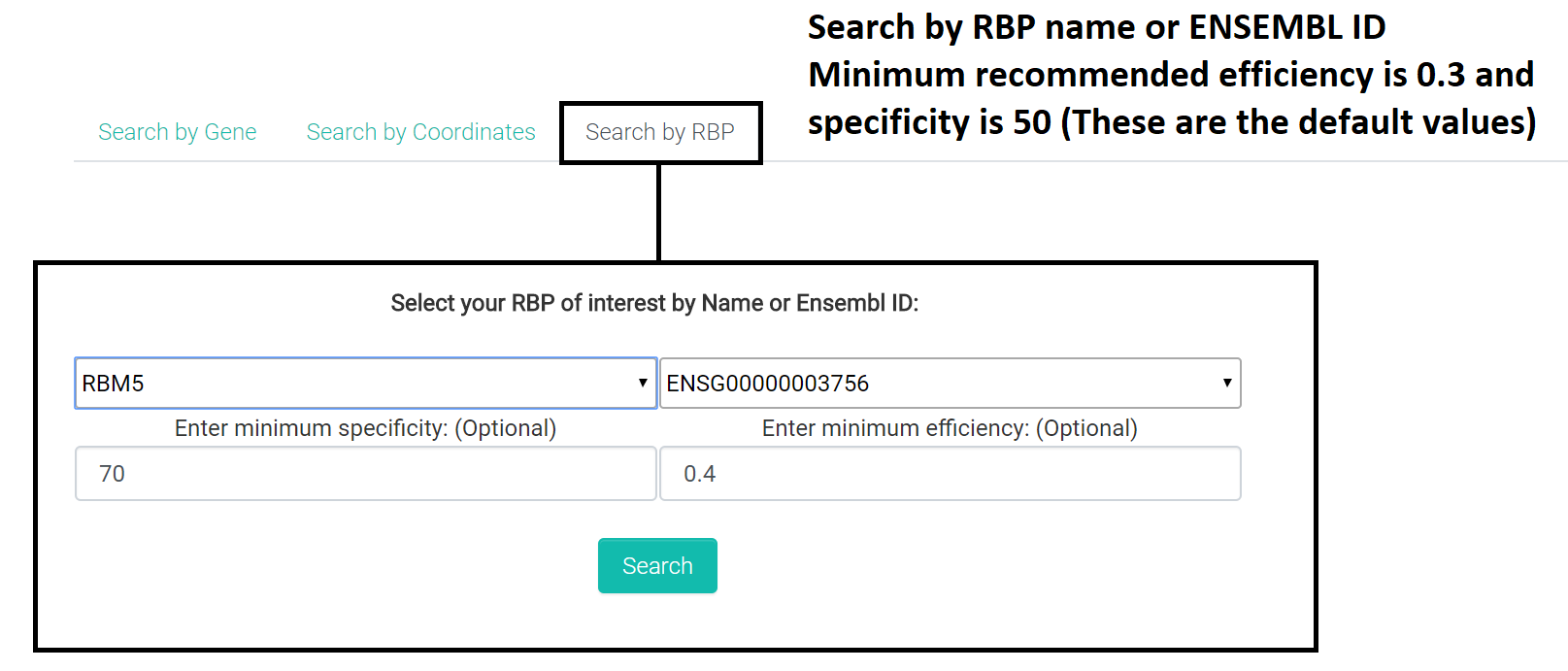
3. RBP based search:
Select the RBP or the corresponding ENSEMBL ID of interest and give a base specificity and efficiency scores.
Default specificity score is 50 and efficiency score is 0.3 and that will be applied to the search if the fields are left empty.
Search results
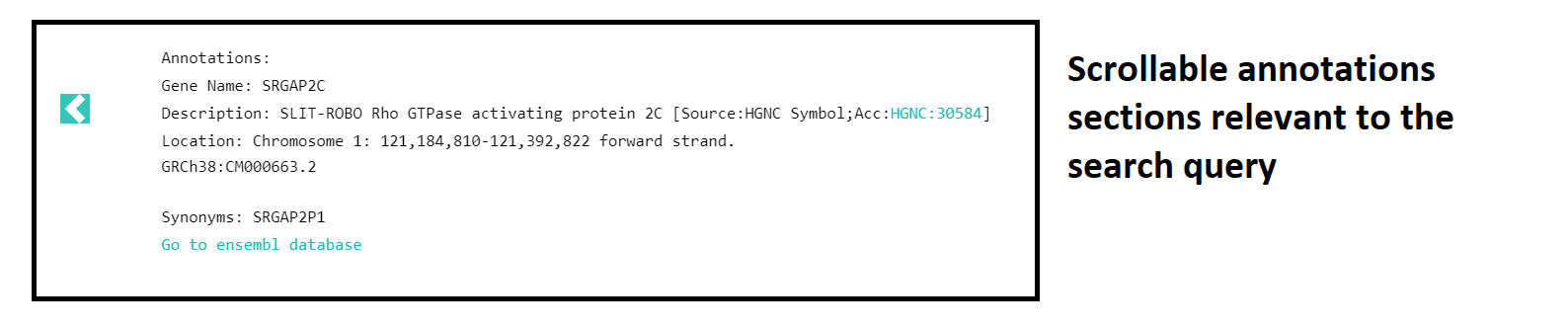
Annotations:
For gene and coordinate based search, scrollable annotations providing more information on search query are displayed on top of the page
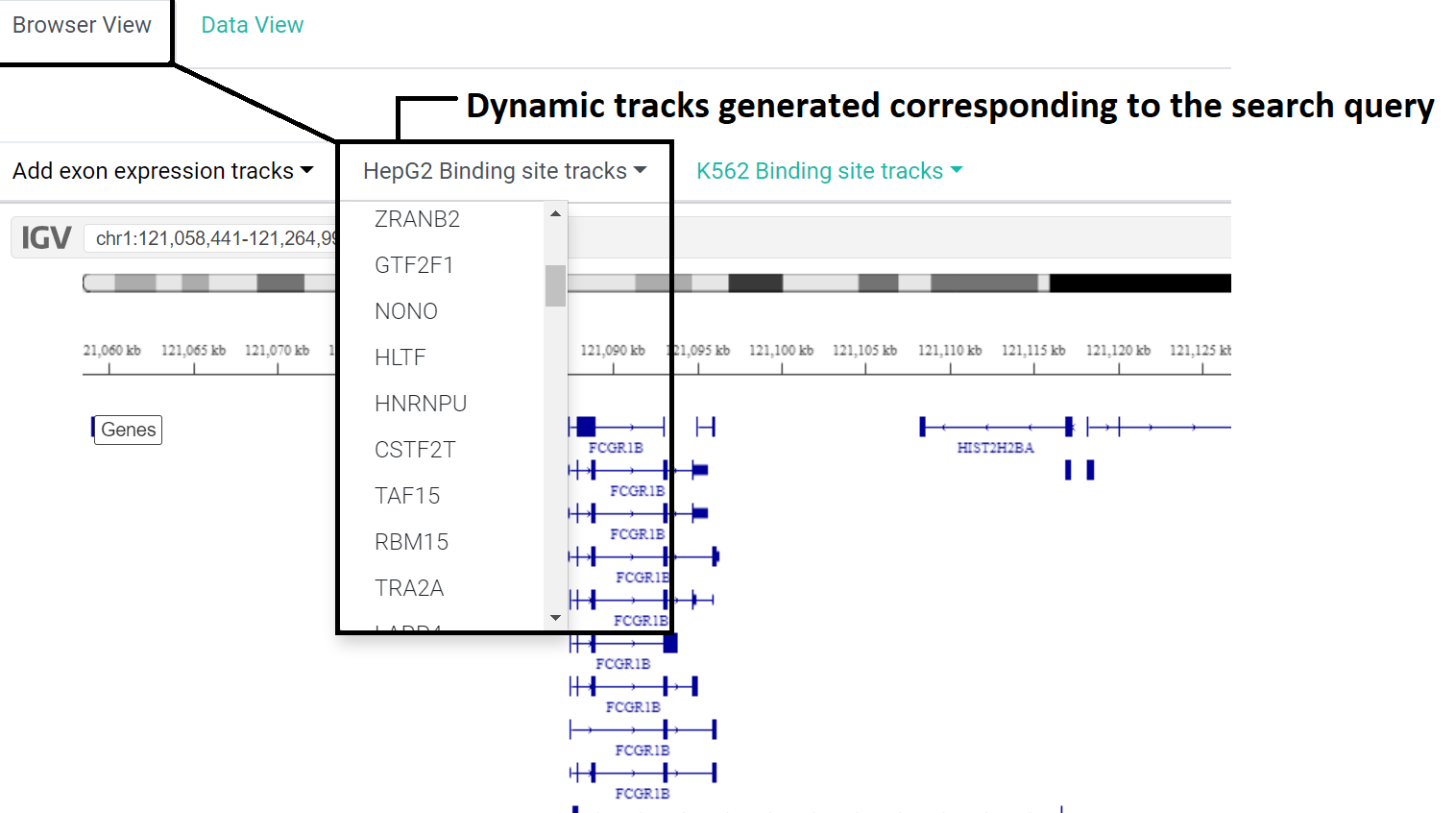
Two types of dynamic tracks generated
1. Premade/already available tracks that are displayed by default
2. Custom tracks corresponding to search query
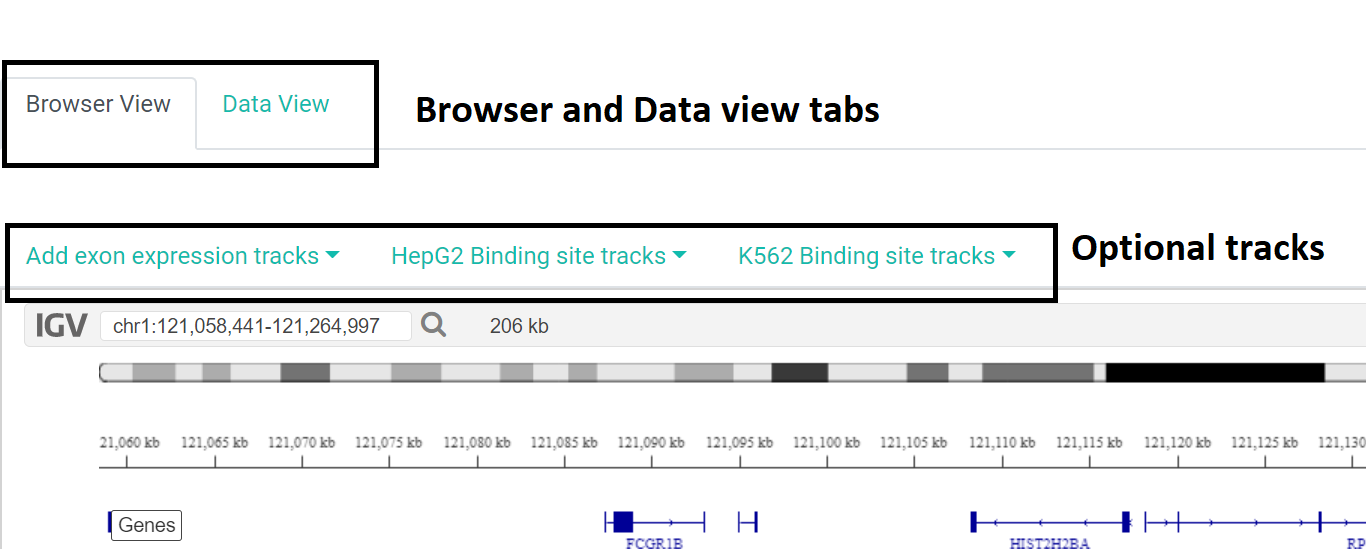
Format 1: Browser view:
This tab is useful for data visualization where the user can compare the results with several other tracks.

Along with the 6 default tracks, the users have the option to choose several optional tracks such as tracks for exon expression across tissues, relevant tracks for specific genes are also generated for both hepg2 and k562 binding sites.
If the search is based on RBP, then an additional optional navigation menu is generated from which user can quickly navigate across various target genes in that network.
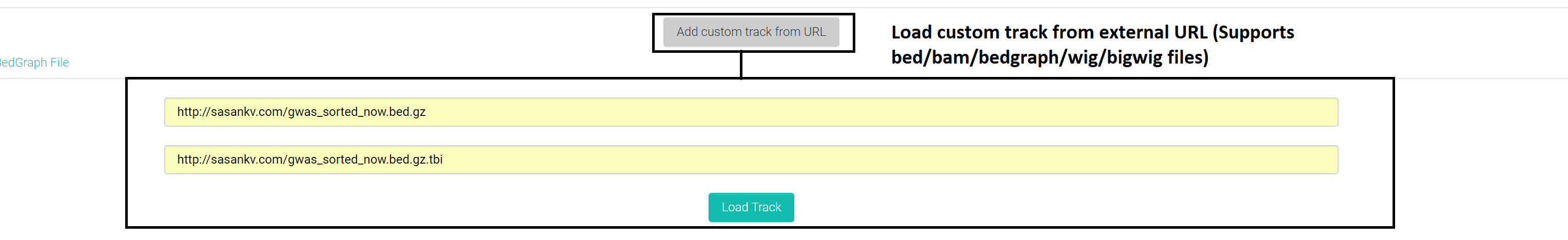
Browser view also lets users add tracks from external URLs and it currently supports BED, BAM, WIG, BIGWIG, BEDGRAPH file formats.
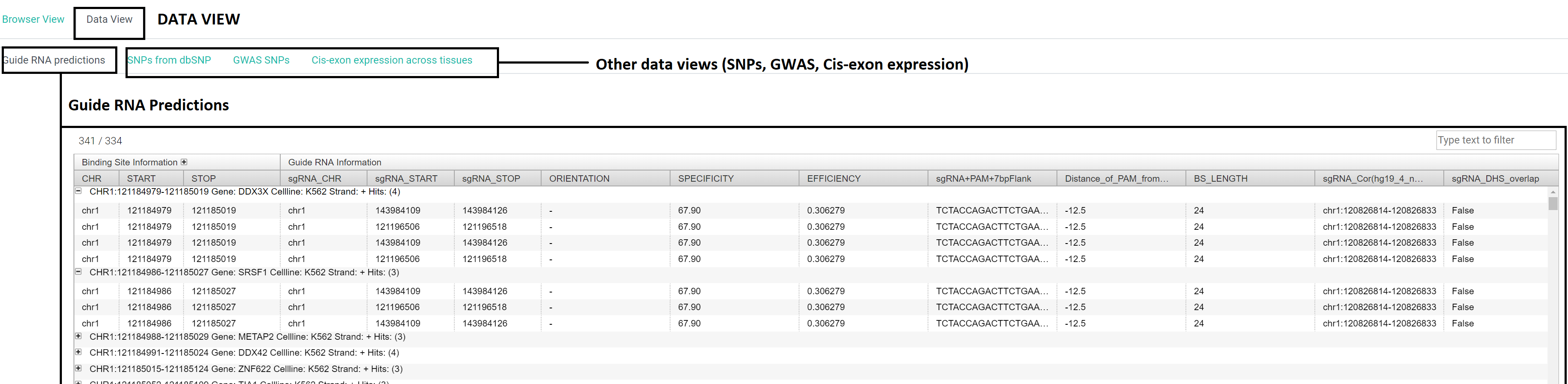
Format 2: Data view:
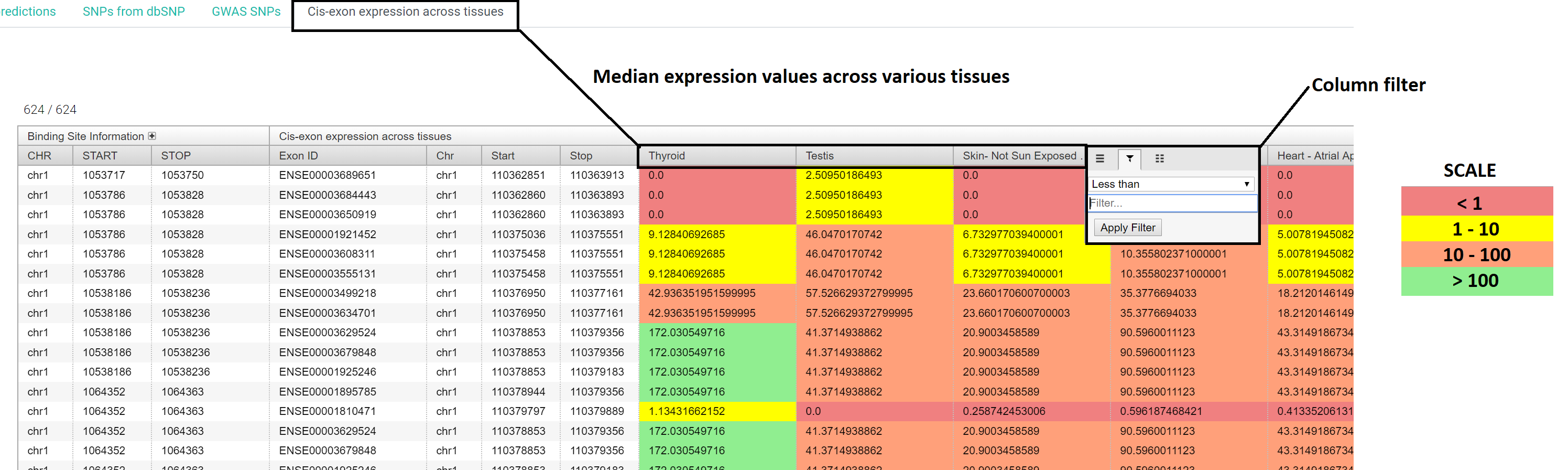
This tab provides raw data output for the search query in form of 4 interactive tables.
The results are binding site centric and the 4 tables are guide RNA predictions for the binding sites, SNPs, GWAS SNPs and proximal exon expression details for the binding sites
The expression values are color coded and the color scale is given below